Abstract
Red wolves are a critically endangered species native to the eastern United States, but despite conservation efforts, their population size has declined and has experienced a decrease in genetic diversity during captive breeding. In recent years, it was discovered that coyotes in the region of Galveston Island retained red wolf alleles lost in the captive-bred population, meaning the two species had hybridized in the past.1 This project analyzed the diet of canids in the Galveston region of Texas and assessed the spatial distribution of the diet components. Canid scat samples were collected around Galveston and analyzed to determine the prey item(s) consumed. The samples were analyzed in ArcGIS Pro to determine location by land cover, in order to better understand the habitats of these canids. The results were of varying success. The most common food items found within the canid samples in this study were chicken and hispid cotton rats. There was no significant relationship between land cover and the prey items consumed. The next step for this project is to analyze the degree of hybridization and maternal haplotype grouping of each canid to determine if prey items consumed vary by those values.
Introduction
Red wolves (Canis rufus) are a critically endangered species endemic to the eastern United States and are now considered extinct in the wild. Captive red wolf populations are small (about 241 individuals) and due to low founder population size, have lost valuable genetic diversity. 2 However, recent years have seen a resurgence of red wolf DNA in an unlikely source: coyotes (Canis latrans). A 2018 study based in Galveston Island in Texas discovered that coyote tissue samples contained red wolf ghost alleles, despite red wolves disappearing from the area since 1980.3 Further studies corroborated this finding and have led to the realization that hybridization took place between red wolves and coyotes prior to their removal from Texas and Louisiana for captive breeding.4


Hybridization refers to the movement of alleles from one species to another. Although it can lead to the mixing between certain species, it can be beneficial in providing a new source of DNA into a species with a slowly dying gene pool.7 The purpose of this study was to continue the work in Galveston County, Texas in order to determine the frequency of these red wolf coyote hybrids and assess the diet of these canids. By discovering how common these hybrids are, potential opportunities for increasing genetic diversity in the captive red wolf population can be assessed. This study had three goals, though not all of them have been completed. The first goal was to analyze canid scat samples and identify the maternal haplotype (mtDNA) level, which indicates whether the canid is a coyote, red wolf, or hybridized coyote. This will provide crucial context for the rest of this project, as understanding this information will grant an opportunity to know the current breakdown of coyote and hybrid individuals in this region.
The second goal of this project was to ascertain whether the canid selected a certain variety of prey items based on mtDNA haplotype group. Coyotes are generalists when it comes to diet. They have a wide variety of food they will consume if the opportunity arises, but it has been found that rodents and rabbits are most common in their diet while other food items include birds, reptiles, and fruits.8 Red wolves share various similarities to the diet of coyotes, as their diet also depends on the availability of prey, but the red wolf diet tends to consist more significantly of rabbits, rodents, raccoons, and white-tailed deer, with the inclusion of a few other small mammals.9 These diet differences could help us identify hybridized coyotes, as it is suspected their larger size due to red wolf ancestry may favor larger prey items like those in the red wolf diet.
The third and final goal was to determine the type of habitat these scat samples were found in and determine whether there was a relationship between land features and prey. According to the U.S. Fish and Wildlife Service, red wolves in southeastern Texas used to be found primarily in “coastal prairie and marsh” habitats, but other habitats within the region that have access to food, water, and shelter, could be home to the red wolf.10 The coyote has a much wider distribution across the United States and can adapt to a variety of habitats. Coyotes prefer natural areas (particularly grasslands and forests) but can adapt to live in urban areas as well.11 I hypothesized that if there was a distinction between the habitats each haplotype preferred, it would be that red wolves were more likely to be found in natural areas while coyotes would be found closer to urban areas.
Methodology
This project used 24 canid scat samples collected around Galveston County, Texas by Dr. Karlin, a professor in the Physics and Environmental Science Department at St. Mary’s University, and her lab assistants. The coordinates of each scat sample location were recorded and imported to ArcGIS Pro. Dr. Karlin analyzed these samples for prey species identification using DNA metabarcoding techniques at Michigan Technological University in May 2022. DNA metabarcoding provided information on the frequency of prey species found in each scat sample. This data was combined with the scat coordinates in ArcGIS Pro and overlaid on a landcover layer for analysis. The land cover data used for this research was the United States Geological Survey (USGS) Multi-Resolution Land Characteristics (MLRC) National Land Cover Dataset (NLCD) from 2015. This dataset sorted land within the United States into 19 different types, including wetlands, mixed forests, and croplands. By analyzing the location of the scat samples within the different land cover types, we could identify if different land cover types had a relationship with prey items or the degree of hybridization of the canids. This latter part of the project is still currently in progress. We used a Spearman’s correlation test to determine if the type of prey item was correlated with land cover type and a one-way ANOVA (Analysis of Variance) test to determine if any prey items were consumed significantly more often than others. Alpha was 0.05 for both statistical tests.
Results
The canid scat samples were found in three land cover types: wetlands, tropical or sub-tropical grassland, and urban and built-up (Figure 1). We are still waiting on haplotype data results, so we were not able to determine the relationship between haplotype and landcover data.
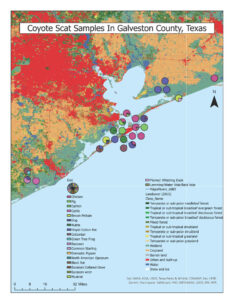
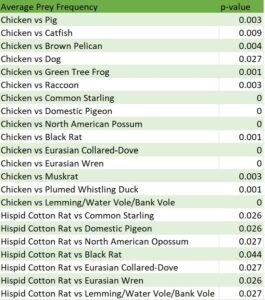
The Spearman correlation tests were not significant for any prey item. The 1-way ANOVA test was significant (F=3.424, df=19, p=<0.001) and the post-hoc comparison identified multiple significant comparisons. Chicken was the prey item most frequently consumed with a mean percentage of 26.54% and the hispid cotton rat was the second most frequently consumed prey item with a mean of 20.50% (Table 1). The prey items with the lowest mean percentage were the common starling, with a mean percentage of 0.02%, and the domestic pigeon, with a mean percentage of 0.03%. The post-hoc comparison results indicated that the significant differences were comparisons with any prey species compared to chicken or hispid cotton rats (Table 2). Chicken had significant relationships with fifteen other prey types while the hispid cotton rat had significant relationships with seven other prey types (Table 2).
Table 1. Mean frequency of each prey item consumed for all scat samples combined.
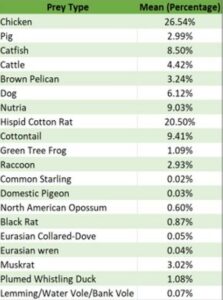
Table 2. Significant post-hoc comparisons by prey species for all scat samples combined.
Discussion
There was no significant relationship between prey items consumed and landcover type, but this may be related to the low differentiation in landcover types using the MLRC NLCD dataset. This dataset generalizes landcover to a 30 m resolution scale and is from 2015 so it is not up to date with the current landcover of the study area. One of the next steps for this goal of the project is to reanalyze the landcover types using a higher resolution and more recent dataset in order to more confidently determine if there is a relationship between prey items and landcover. Additionally, once the haplotype of each sample is established, we will be able to determine whether a prey item is more commonly consumed by coyotes or hybrids. For the time being, I can determine that chicken was most often consumed often by the canids, especially compared to the other prey items within this study. The same is true for the hispid cotton rat, though to a lesser extent as it was the second most eaten prey item and had significant relationships with seven other prey item categories. An interesting next step with this dataset would be to try and determine the source of chicken – people often keep chickens for egg or meat production, but because Galveston is highly urbanized on the east end, there may also be significant access to garbage or dump sites where these canids could be scavenging.



- Heppenheimer, E., Brzeski, K., Wooten, R., Waddell, W., Rutledge, L., Chamberlain, M., Stahler, D., Hinton, J., & vonHoldt, B. (2018). Rediscovery of Red Wolf Ghost Alleles in a Canid Population Along the American Gulf Coast. Genes, 9(12), 618. https://doi.org/10.3390/genes9120618 ↵
- Red Wolf (Canis rufus) | U.S. Fish & Wildlife Service. N.d. FWS.gov. https://www.fws.gov/species/red-wolf-canis-rufus. ↵
- Heppenheimer, E., Brzeski, K., Wooten, R., Waddell, W., Rutledge, L., Chamberlain, M., Stahler, D., Hinton, J., & vonHoldt, B. (2018). Rediscovery of Red Wolf Ghost Alleles in a Canid Population Along the American Gulf Coast. Genes, 9(12), 618. https://doi.org/10.3390/genes9120618 ↵
- Barnes, T. M., Karlin, M., vonHoldt, B. M., Adams, J. R., Waits, L. P., Hinton, J. W., Henderson, J., & Brzeski, K. E. (2022). Genetic diversity and family groups detected in a coyote population with red wolf ancestry on Galveston Island, Texas. BMC Ecology & Evolution, 22(1), 1–14. https://doi.org/10.1186/s12862-022-02084-9 ↵
- Wolf, Red. 2011. Female Red Wolf in the Snow. Photo. https://www.flickr.com/photos/trackthepack/6189661866/. ↵
- Couperus, Jitze. 2007. Coyote. Photo. https://www.flickr.com/photos/jitze1942/1751241022/. ↵
- Barnes, T. M., Karlin, M., vonHoldt, B. M., Adams, J. R., Waits, L. P., Hinton, J. W., Henderson, J., & Brzeski, K. E. (2022). Genetic diversity and family groups detected in a coyote population with red wolf ancestry on Galveston Island, Texas. BMC Ecology & Evolution, 22(1), 1–14. https://doi.org/10.1186/s12862-022-02084-9 ↵
- S.A. Poessel, E.C. Mock, and S.W. Breck. 2017. Coyote (Canis latrans) diet in an urban environment: variation relative to pet conflicts, housing density, and season. Canadian Journal of Zoology. 95(4): 287-297. https://doi.org/10.1139/cjz-2016-0029. ↵
- Red Wolf (Canis rufus) | U.S. Fish & Wildlife Service. N.d. FWS.gov. https://www.fws.gov/species/red-wolf-canis-rufus. ↵
- Red Wolf (Canis rufus) | U.S. Fish & Wildlife Service. N.d. FWS.gov. https://www.fws.gov/species/red-wolf-canis-rufus. ↵
- Franckowiak GA, Perdicas M, Smith GA. 2019. Spatial ecology of coyotes in the urbanizing landscape of the Cuyahoga Valley, Ohio. PLoS One. 14(12):e0227028. doi:10.1371/journal.pone.0227028. ↵
- AnemoneProjectors. 2009. Buck Buck Buck. Chicken Uploaded by MaybeMaybeMaybe. https://commons.wikimedia.org/wiki/File:Chicken_free_range_1.jpg. ↵
- J. N. Stuart. 2011. Hispid Cotton Rat (Sigmodon Hispidus). Photo. https://www.flickr.com/photos/stuartwildlife/5727265600/. ↵
- ucumari photography. 2009. Red Wolf (Canis Rufus). Photo. https://www.flickr.com/photos/ucumari/4237167628/. ↵




4 comments
Andrew Ponce
Hello Bianca. This article is certainly one of the most interesting on this site. Understanding the different situations in which animals change, play a huge role in understanding how the balance of nature works in relation to coyotes and wolves. This article brings light to a subject that not many people have even thought of. The background of animals, the resemblance of some, and so many more facts. Very informative, and very interesting. Great job!
Gaitan Martinez
Wolves are honestly my favorite animal, but I will admit, I don’t know much about them. So this article really pushes me to want to research more about it. I do know some info about other animals, but not wolves. Great article, didn’t even know red wolves existed, and you can see the resemblance from both the fox and wolf.
Jared Sherer
After reading your article I got to learn a lot about red wolves and how they function. Your article was well written and very informative. I enjoyed that it was easy to follow and I appreciate the data that was provided in the text. I got to learn about this species for the first time which I found fun to read about. Awesome job!
Matthew Holland
It is one of the best of its kind in it the respective section hands down. I look forward to reading more from them in the future and hope they can write about different topics that would be equally interesting. I am looking forward to hearing more about how the situations with these animals change as it is extremely important to keep nature in balance with animals such as wolves and coyotes.